Featured Tools
Data Explorer
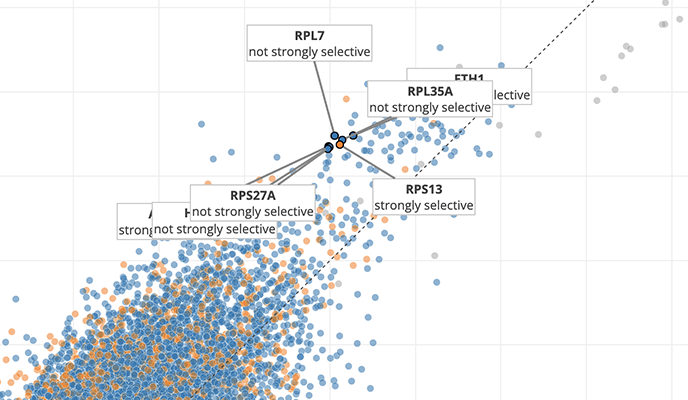
Use the Data Explorer tool to dive deeper into DepMap datasets and explore relationships across cell lines.
Cell Line Selector

Need help selecting cell lines? Use the Cell Line Selector to explore and choose lines of interest and and build customized cell line lists.
Data Downloads
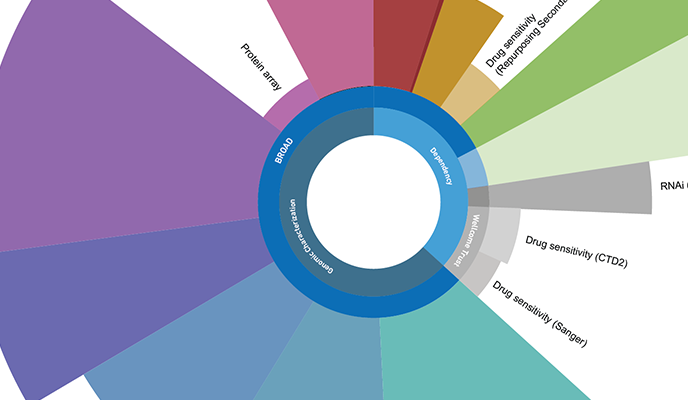
Browse and access the complete collection of datasets available in the DepMap portal, including the latest released data.
Recent Highlights
View the latest DepMap announcements and release notes
| 10.01.25 |
The 25Q3 release is now live and contains new CRISPR screens and Omics data, including more data from the Pediatric Cancer Dependencies Accelerator, and a new tool, Context Explorer, that allows users to explore enriched genetic dependencies and compound sensitivities across lineage and subtype contexts. Please read the release notes for more details. |
| 06.27.25 |
The 25Q2 release is now live and contains new data, including new data from the Pediatric Cancer Dependencies Accelerator, pipeline improvements, updates to citing DepMap data and more! Please read the release notes for more details. |
| 12.16.24 |
The 24Q4 data and portal updates are now live! Please read the release notes for more details. |
| 06.03.24 |
The 24Q2 release is now live and contains new cell lines and datasets, metadata and pipeline improvements. Please read the release notes for more details. |
| 01.08.24 |
Fixed bug which caused some compound names to not show data from GDSC screens |
| 11.27.23 |
The 23Q4 release is now live and contains new data, updates to portal tools, metadata and pipeline improvements. Please read the release notes for more details. |
| 07.10.23 |
Bug fix: The "Peturbation Effects" tab of the gene page was not correctly coloring points for lines with hotspot or damaging mutations. This has been fixed. |
| 07.06.23 |
We've added additional single point compound screening data from the PRISM Repurposing project to the portal. You can see it in the portal labeled, "PRISM Repurposing Public 23Q2" and is a superset of the data that was previously released as "Repurposing Primary" |
| 05.09.23 |
We are excited to share new CRISPR and omics data, a Chronos update and a new Target Discovery app in this 23Q2 release! You’ll also notice an update to our cell line page. We hope this new format will allow users to more easily navigate the information presented for each cell line. In this release, you will likely notice some changes in the data now that Chronos 2.0 integrated CRISPR data is being produced by a joint Chronos run. This change reduces the number of false positives in the integrated dataset by 5%. Please read the full announcement for more information. Additionally, we are aware that there is an artifact in the CRISPR data which causes background correlation in dependency between genes located on the same chromosome arm. To account for this, we've aligned the mean gene effect of each chromosome arm to be the same in every cell line following the original copy number correction. Overall we see an improvement in data quality, as well as a reduction in clustering by chromosome arm in UMAP embeddings. See more information about this release see this forum post describing the full set of changes. |
View All Announcements
Community Forum
Have questions, comments, or feedback?
The DepMap Community Forum is the space for you to ask questions, share your suggestions, and report issues that the community can investigate and solve.
Visit Q&AFollow us for news and updates
Stay up-to-date on our latest tools and releases by following us on LinkedIn and X(Twitter).
Explore More Ways To Use DepMap
Discover genetic and pharmacological dependencies
Explore MSI cancer dependency on WRN in Data ExplorerSee the Gene Dependency Overview page for Sox10
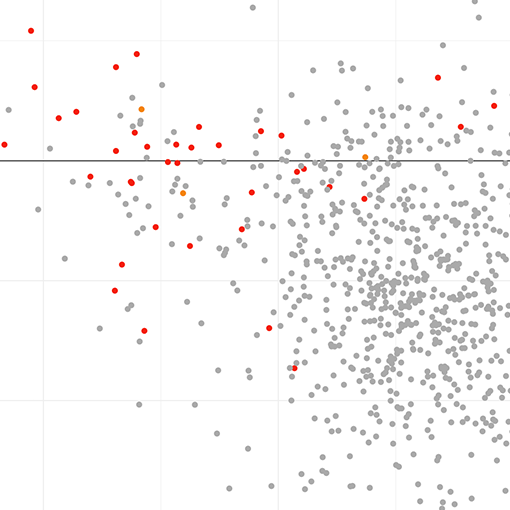
Prioritize tumor contexts and predictive biomarkers
Compare TRILACICLIB (CDK inhibitor) and RB1 expression in Data Explorer
Browse our collection of over 2000 cancer models
Dive deeper into MCF7 on our Gene Dependency page
Discover interesting targets
Find the most selective and/or predictable gene dependencies in Target discoveryContribute Cancer Models and Datasets to DepMap
